Segmentation of organs and tumors from medical images, like computed tomography (CT) and magnetic resonance (MR) imaging, are critical for radiation therapy planning, image-guided surgery, disease diagnois and functionality assessment. Manual delineation of organs and tumors from hundreds of CT and MR slices per scan is labor and time consuming. Traditional computer vision and image processing methods are not robust given the variations in image quality (spatial resolutions, signal-to-noise ratio) and human anatomy, and can take a significant amount of time (in minutes) and sometimes require user interactive to provide additional input.
A deep learning model taught to learn from the variations in medical image quality and human anatomy by training on a large dataset size. The segmentation predictions are also fully automated, fast (in seconds) and require no user intervention.
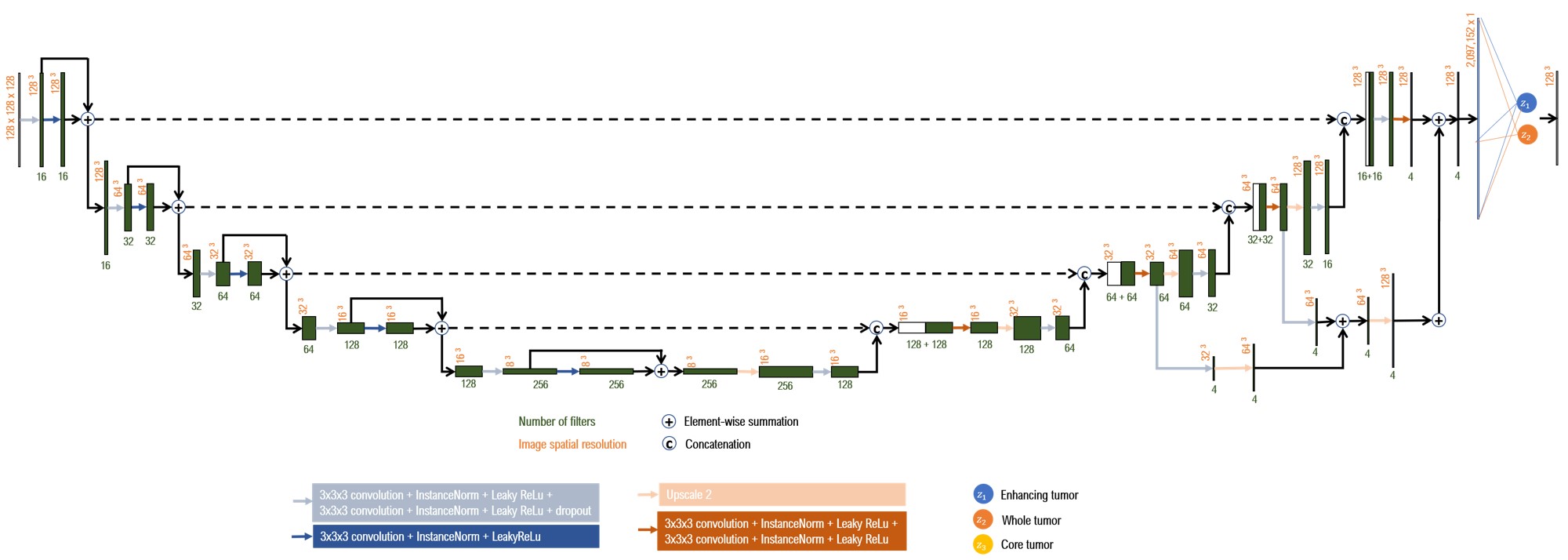
3D U-Net architecture
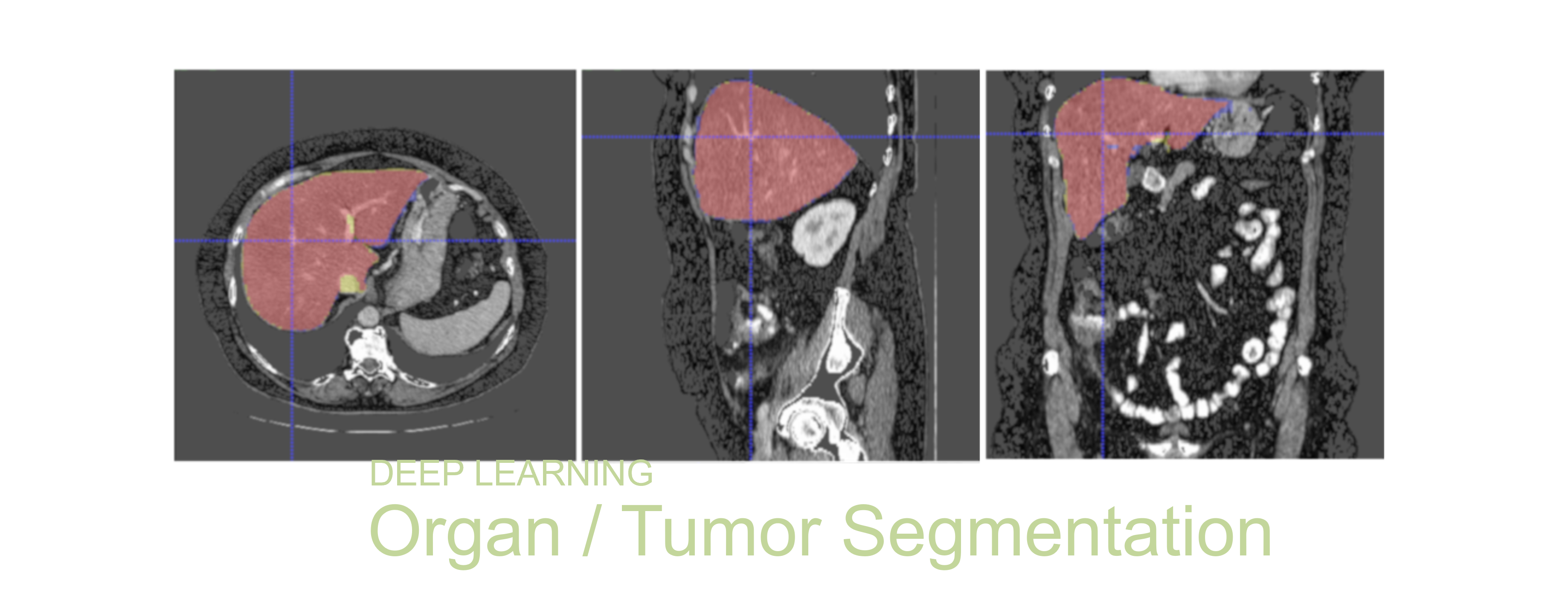
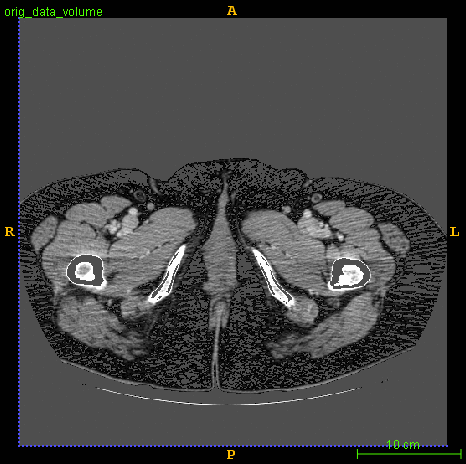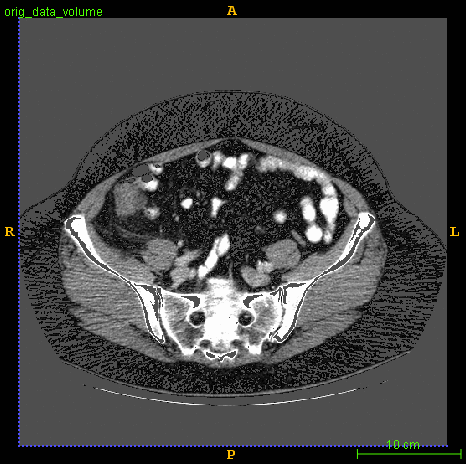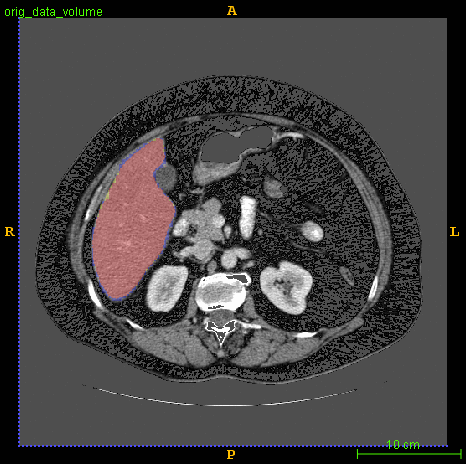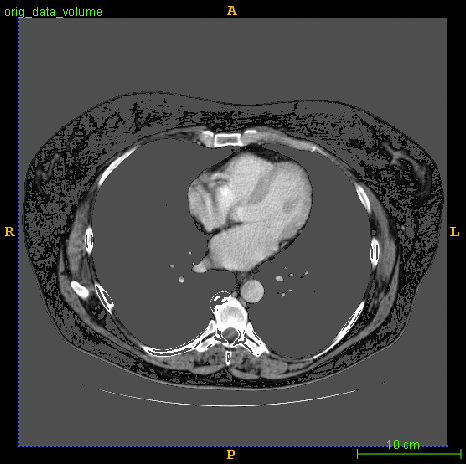
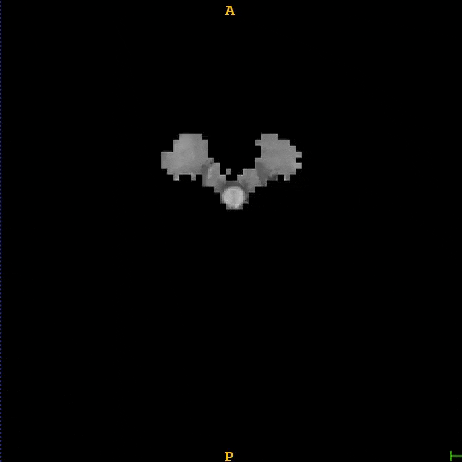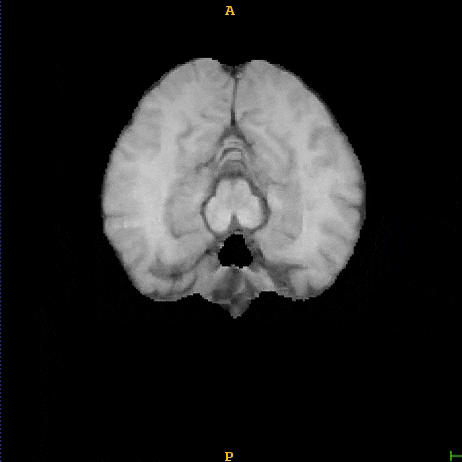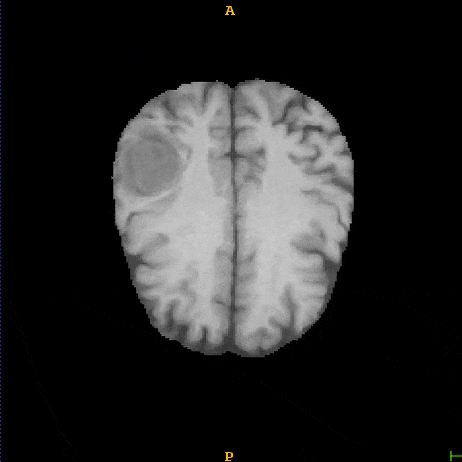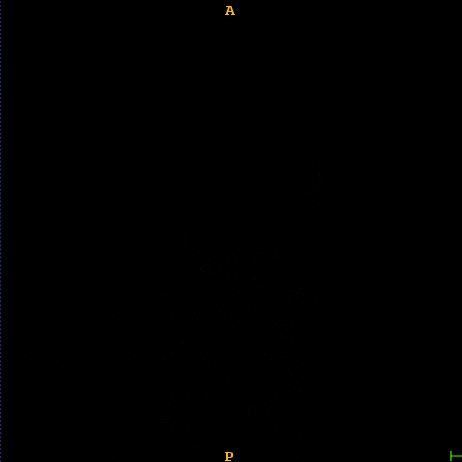
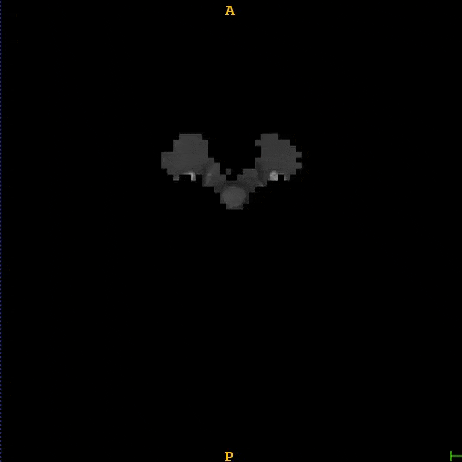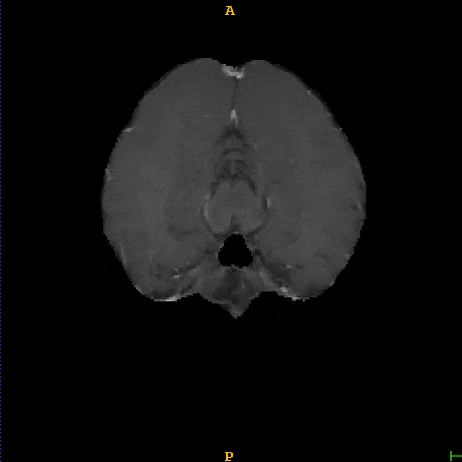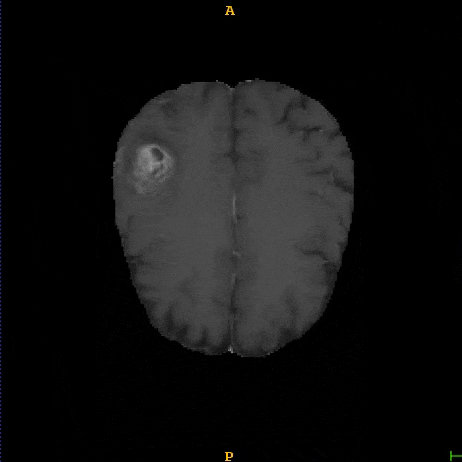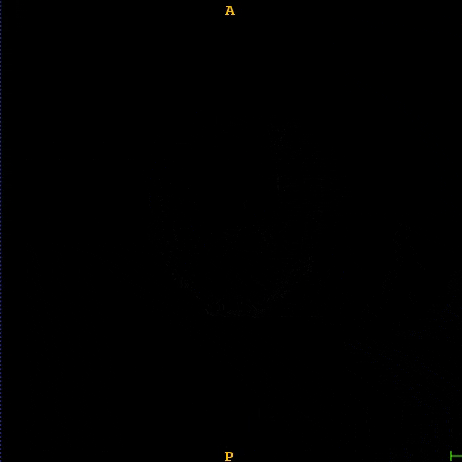
Segmented liver organ from abdominal CT predicted using deep learning
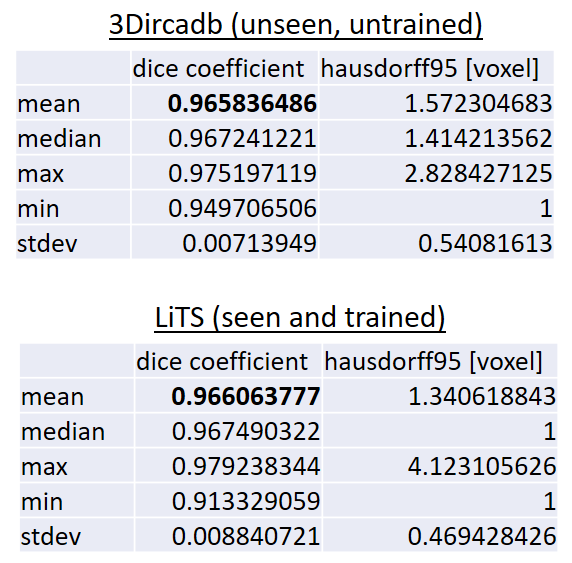
A 3D U-Net based deep learning model was developed and trained with segmented liver organs from CTs from the Liver Tumor Segmentation Challenge (LiTS) . A different and unseen dataset from 3Dircadb was used for independent validation. High dice and Hausdorff 95 scores were observed for the predicted liver segmentations.
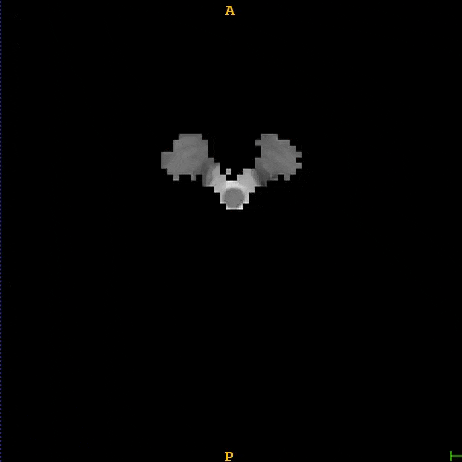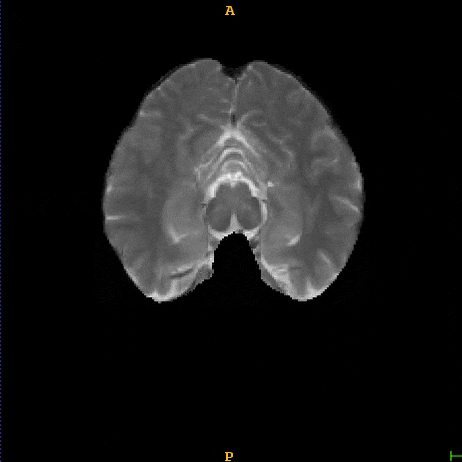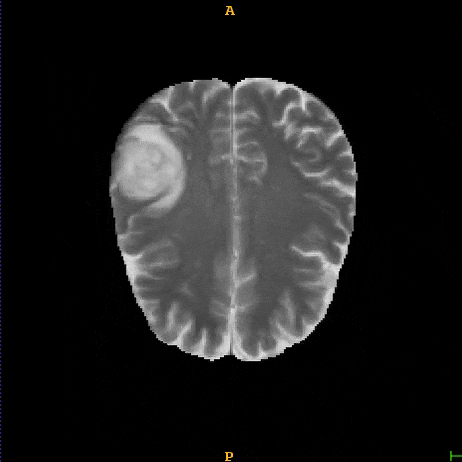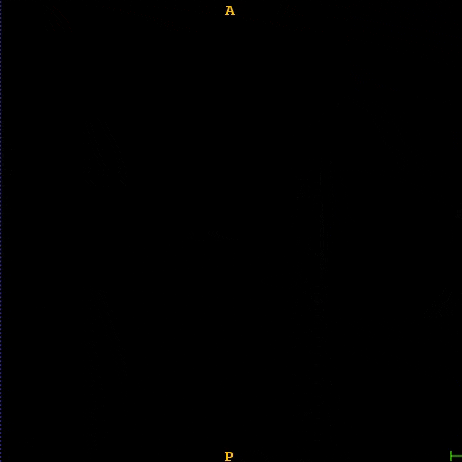
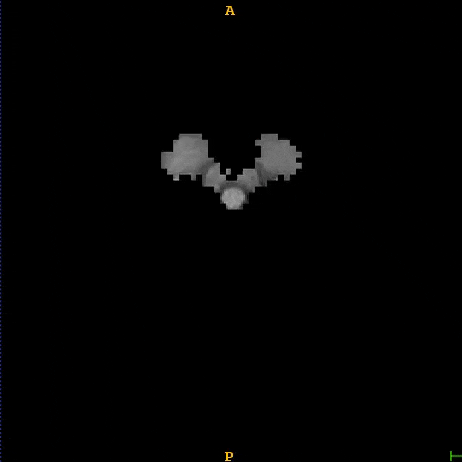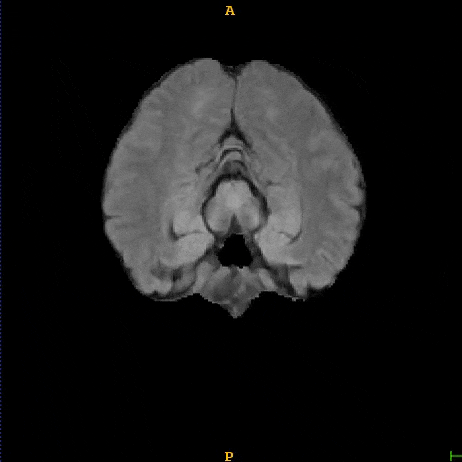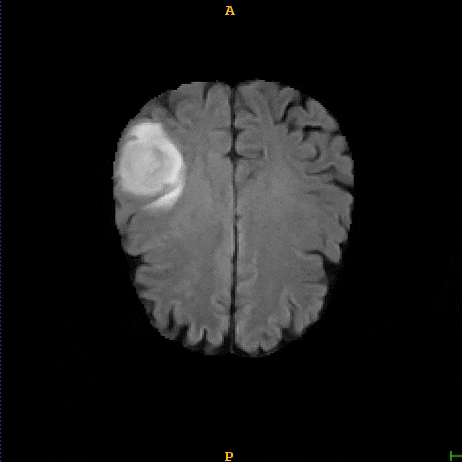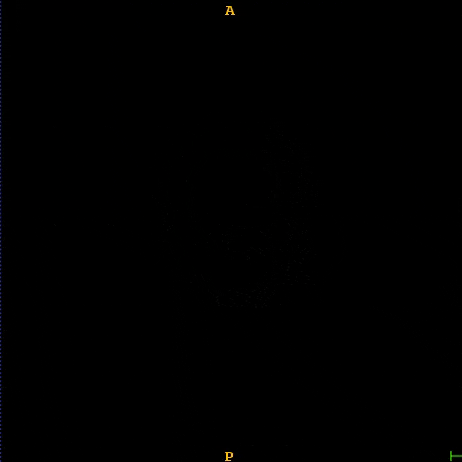
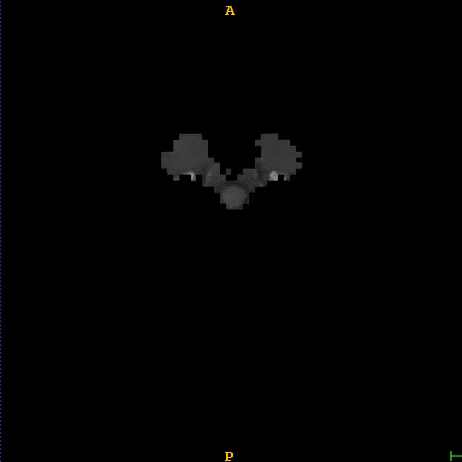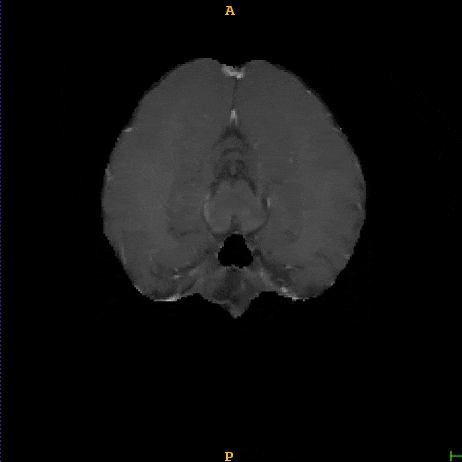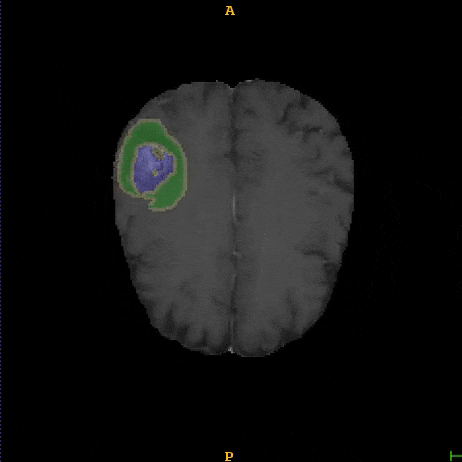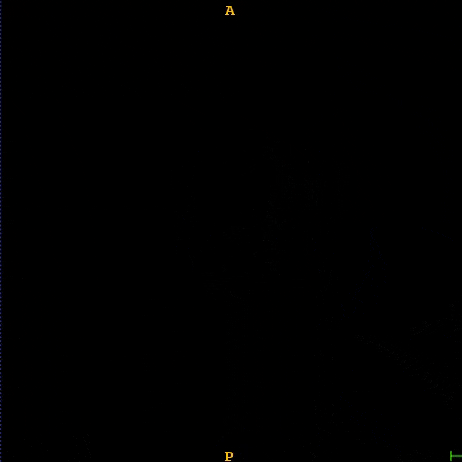
Segmented brain tumor from MR predicted using deep learning
A similar 3D U-Net architecture was also applied towards multiclass segmentation of brain tumor from MRIs provided by the Multimodal Brain Tumor Segmentation Challenge 2018 (BraTS) . High dice and Hausdorff 95 scores were observed for the predicted brain tumor segmentations.
Contact us for more information.